This page was built from the example_notebook Jupyter notebook available on Github
How to use¶
These pages will outline basic usage of DefDAP, including loading a DIC and EBSD map, linking them with homologous points and producing maps
Load in packages¶
DefDAP is split into modules for processing EBSD (defdap.ebsd) and HRDIC (defdap.hrdic) data. There are also modules for manpulating orientations (defdap.quat) and creating custom figures (defdap.plotting) which is introduced later. We also import some of the usual suspects of the python scientific stack: numpy and matplotlib.
[1]:
import numpy as np
import matplotlib.pyplot as plt
import defdap.hrdic as hrdic
import defdap.ebsd as ebsd
from defdap.quat import Quat
# try tk, qt, osx (if using mac) or notebook for interactive plots. If none work, use inline
%matplotlib inline
Load in a HRDIC map¶
[2]:
dic_filepath = "../../tests/data/testDataDIC.txt"
dic_map = hrdic.Map(dic_filepath)
Starting loading HRDIC data.. Loaded DaVis 8.4.0 data (dimensions: 300 x 300 pixels, sub-window size: 12 x 12 pixels)
</pre>
Starting loading HRDIC data.. Loaded DaVis 8.4.0 data (dimensions: 300 x 300 pixels, sub-window size: 12 x 12 pixels)
end{sphinxVerbatim}
Starting loading HRDIC data.. Loaded DaVis 8.4.0 data (dimensions: 300 x 300 pixels, sub-window size: 12 x 12 pixels)
Set the scale of the map¶
This is defined as the pixel size in the DIC pattern images, measured in microns per pixel.
[3]:
field_width = 20 # microns
num_pixels = 2048
dic_map.set_scale(field_width / num_pixels)
Plot the map with a scale bar¶
[4]:
dic_map.plot_map('max_shear', vmin=0, vmax=0.10, plot_scale_bar=True)
[4]:
<defdap.plotting.MapPlot at 0x7fe47f92faf0>
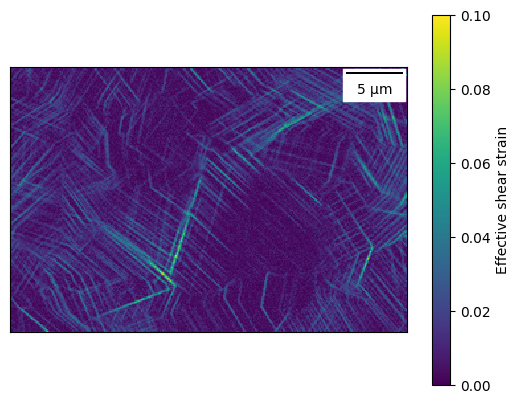
Crop the map¶
HRDIC maps often contain spurious data at the edges which should be removed before performing any analysis. The crop is defined by the number of points to remove from each edge of the map, where xMin, xMax, yMin and yMax are the left, right, top and bottom edges respectively. Note that the test data doesn not require cropping as it is a subset of a larger dataset.
[5]:
dic_map.set_crop(left=0, right=0, top=0, bottom=0)
Statistics¶
Some simple statistics such as the minimum, mean and maximum of the effective shear strain, E11 and E22 components can be printed.
[6]:
dic_map.print_stats_table(percentiles=[0, 50, 100], components = ['max_shear', 'e'])
dic_map (dimensions: 300 x 200 pixels, sub-window size: 12 x 12 pixels, number of points: 60000)
Component 0 50 100
max_shear 0.0000 0.0085 0.0910
e11 -0.1083 -0.0097 0.0168
e12 -0.0478 0.0005 0.0630
e21 -0.0478 0.0005 0.0630
e22 -0.0493 0.0045 0.0850
Set the location of the DIC pattern images¶
The pattern images are used later to define the position of homologous material points. The path is relative to the directory set when loading in the map. The second parameter is the pixel binning factor of the image relative to the DIC sub-region size i.e. the number of pixels in the image across a single datapoint in the DIC map. We recommend binning the pattern images by the same factor as the DIC sub-region size, doing so enhances the contrast between microstructure features.
[7]:
# set the path of the pattern image, this is relative to the location of the DIC data file
dic_map.set_pattern("testDataPat.bmp", 1)
Load in an EBSD map¶
Currently, OxfordBinary (a .crc and .cpr file pair), OxfordText (.ctf file), EdaxAng (.ang file) or PythonDict (Python dictionary) filetypes are supported. The crystal structure and slip systems are automatically loaded for each phase in the map. The orientation in the EBSD are converted to a quaternion representation so calculations can be applied later.
[8]:
ebsd_map = ebsd.Map("../../tests/data/testDataEBSD.cpr")
Starting loading EBSD data.. Loaded EBSD data (dimensions: 359 x 243 pixels, step size: 0.12 um)
</pre>
Starting loading EBSD data.. Loaded EBSD data (dimensions: 359 x 243 pixels, step size: 0.12 um)
end{sphinxVerbatim}
Starting loading EBSD data.. Loaded EBSD data (dimensions: 359 x 243 pixels, step size: 0.12 um)
A list of detected phases and crystal structures can be printed
[9]:
for i, phase in enumerate(ebsd_map.phases):
print(i+1)
print(phase)
1
Phase: Ni-superalloy
Crystal structure: cubic
Lattice params: (3.57, 3.57, 3.57, 90, 90, 90)
Slip systems: cubic_fcc
A list of the slip planes, colours and slip directions can be printed
[10]:
ebsd_map.phases[0].print_slip_systems()
Plane 0: (111) Colour: blue
Direction 0: [011̅]
Direction 1: [1̅01]
Direction 2: [11̅0]
Plane 1: (111̅) Colour: green
Direction 0: [011]
Direction 1: [1̅01̅]
Direction 2: [11̅0]
Plane 2: (1̅11) Colour: red
Direction 0: [011̅]
Direction 1: [101]
Direction 2: [1̅1̅0]
Plane 3: (11̅1) Colour: white
Direction 0: [01̅1̅]
Direction 1: [1̅01]
Direction 2: [110]
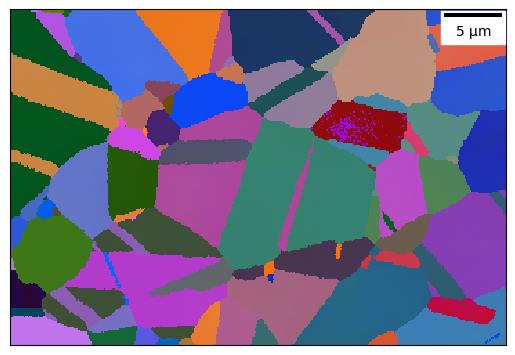
Plot the EBSD map¶
Using an Euler colour mapping or inverse pole figure colouring with the sample reference direction passed as a vector.
[11]:
ebsd_map.plot_map('euler_angle', 'all_euler', plot_scale_bar=True)
[11]:
<defdap.plotting.MapPlot at 0x7fe47f7c5d80>
[12]:
ebsd_map.plot_map('orientation', 'IPF_x', plot_scale_bar=True)
- Starting building quaternion array..
</pre>
- Starting building quaternion array..
end{sphinxVerbatim}
Starting building quaternion array..
Starting building quaternion array.. 100 % Finished building quaternion array (0:00:00)
</pre>
Starting building quaternion array.. 100 % Finished building quaternion array (0:00:00)
end{sphinxVerbatim}
Starting building quaternion array.. 100 % Finished building quaternion array (0:00:00)
[12]:
<defdap.plotting.MapPlot at 0x7fe47f7c5e10>

A KAM map can also be plotted as follows
[13]:
ebsd_map.plot_map('KAM', vmin=0, vmax=2*np.pi/180)
Starting calculating KAM.. Starting calculating KAM.. 100 % Finished calculating KAM (0:00:00)
</pre>
Starting calculating KAM.. Starting calculating KAM.. 100 % Finished calculating KAM (0:00:00)
end{sphinxVerbatim}
Starting calculating KAM.. Starting calculating KAM.. 100 % Finished calculating KAM (0:00:00)
[13]:
<defdap.plotting.MapPlot at 0x7fe47f78d120>
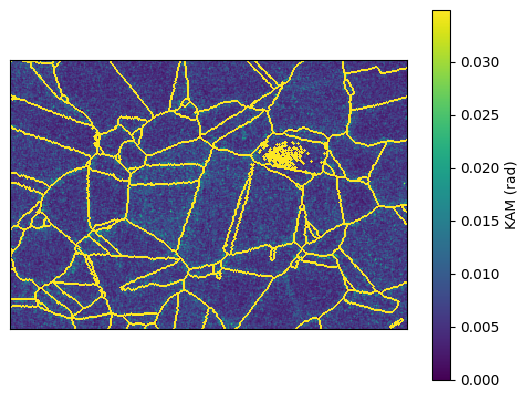
Detect grains in the EBSD¶
This is done in two stages: first bounaries are detected in the map as any point with a misorientation to a neighbouring point greater than a critical value (boundDef in degrees). A flood fill type algorithm is then applied to segment the map into grains, with any grains containining fewer than a critical number of pixels removed (minGrainSize in pixels). The data e.g. orientations associated with each grain are then stored (referenced strictly, the data isn’t stored twice) in a grain
object and a list of the grains is stored in the EBSD map (named grainList). This allows analysis routines to be applied to each grain in a map in turn.
[14]:
ebsd_map.data.generate('grain_boundaries', misori_tol=8)
ebsd_map.data.generate('grains', min_grain_size=10)
- Starting finding grain boundaries..
</pre>
- Starting finding grain boundaries..
end{sphinxVerbatim}
Starting finding grain boundaries..
Starting finding grain boundaries.. 100 % Finished finding grain boundaries (0:00:00)
- Starting finding grains..
</pre>
Starting finding grain boundaries.. 100 % Finished finding grain boundaries (0:00:00)
- Starting finding grains..
end{sphinxVerbatim}
Starting finding grain boundaries.. 100 % Finished finding grain boundaries (0:00:00)
Starting finding grains..
Starting finding grains.. 86 % Finished finding grains (0:00:00)
</pre>
Starting finding grains.. 86 % Finished finding grains (0:00:00)
end{sphinxVerbatim}
Starting finding grains.. 86 % Finished finding grains (0:00:00)
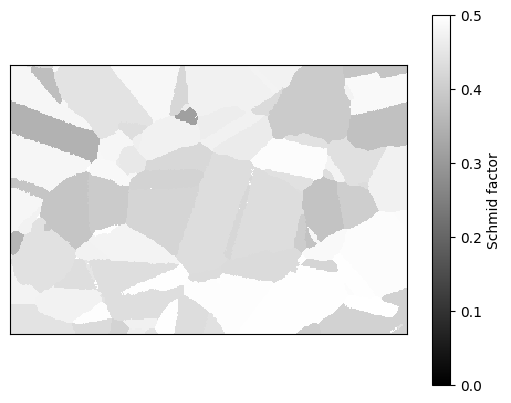
The Schmid factors for each grain can be calculated and plotted. The slipSystems argument can be specified, to only calculate the Schmid factor for certain planes, otherwise the maximum for all slip systems is calculated.
[15]:
ebsd_map.calc_average_grain_schmid_factors(load_vector=np.array([1,0,0]), slip_systems=None)
Starting calculating grain average Schmid factors.. Starting calculating grain average Schmid factors.. 2 % Starting calculating grain average Schmid factors.. 4 % Starting calculating grain average Schmid factors.. 6 % Starting calculating grain average Schmid factors.. 8 % Starting calculating grain average Schmid factors.. 9 %
</pre>
Starting calculating grain average Schmid factors.. Starting calculating grain average Schmid factors.. 2 % Starting calculating grain average Schmid factors.. 4 % Starting calculating grain average Schmid factors.. 6 % Starting calculating grain average Schmid factors.. 8 % Starting calculating grain average Schmid factors.. 9 %
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. Starting calculating grain average Schmid factors.. 2 % Starting calculating grain average Schmid factors.. 4 % Starting calculating grain average Schmid factors.. 6 % Starting calculating grain average Schmid factors.. 8 % Starting calculating grain average Schmid factors.. 9 %
Starting calculating grain average Schmid factors.. 11 % Starting calculating grain average Schmid factors.. 13 % Starting calculating grain average Schmid factors.. 15 % Starting calculating grain average Schmid factors.. 17 % Starting calculating grain average Schmid factors.. 19 % Starting calculating grain average Schmid factors.. 21 % Starting calculating grain average Schmid factors.. 23 % Starting calculating grain average Schmid factors.. 25 % Starting calculating grain average Schmid factors.. 26 %
</pre>
Starting calculating grain average Schmid factors.. 11 % Starting calculating grain average Schmid factors.. 13 % Starting calculating grain average Schmid factors.. 15 % Starting calculating grain average Schmid factors.. 17 % Starting calculating grain average Schmid factors.. 19 % Starting calculating grain average Schmid factors.. 21 % Starting calculating grain average Schmid factors.. 23 % Starting calculating grain average Schmid factors.. 25 % Starting calculating grain average Schmid factors.. 26 %
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. 11 % Starting calculating grain average Schmid factors.. 13 % Starting calculating grain average Schmid factors.. 15 % Starting calculating grain average Schmid factors.. 17 % Starting calculating grain average Schmid factors.. 19 % Starting calculating grain average Schmid factors.. 21 % Starting calculating grain average Schmid factors.. 23 % Starting calculating grain average Schmid factors.. 25 % Starting calculating grain average Schmid factors.. 26 %
Starting calculating grain average Schmid factors.. 28 % Starting calculating grain average Schmid factors.. 30 % Starting calculating grain average Schmid factors.. 32 % Starting calculating grain average Schmid factors.. 34 % Starting calculating grain average Schmid factors.. 36 % Starting calculating grain average Schmid factors.. 38 % Starting calculating grain average Schmid factors.. 40 %
</pre>
Starting calculating grain average Schmid factors.. 28 % Starting calculating grain average Schmid factors.. 30 % Starting calculating grain average Schmid factors.. 32 % Starting calculating grain average Schmid factors.. 34 % Starting calculating grain average Schmid factors.. 36 % Starting calculating grain average Schmid factors.. 38 % Starting calculating grain average Schmid factors.. 40 %
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. 28 % Starting calculating grain average Schmid factors.. 30 % Starting calculating grain average Schmid factors.. 32 % Starting calculating grain average Schmid factors.. 34 % Starting calculating grain average Schmid factors.. 36 % Starting calculating grain average Schmid factors.. 38 % Starting calculating grain average Schmid factors.. 40 %
Starting calculating grain average Schmid factors.. 42 % Starting calculating grain average Schmid factors.. 43 % Starting calculating grain average Schmid factors.. 45 % Starting calculating grain average Schmid factors.. 47 % Starting calculating grain average Schmid factors.. 49 % Starting calculating grain average Schmid factors.. 51 % Starting calculating grain average Schmid factors.. 53 % Starting calculating grain average Schmid factors.. 55 % Starting calculating grain average Schmid factors.. 57 % Starting calculating grain average Schmid factors.. 58 % Starting calculating grain average Schmid factors.. 60 %
</pre>
Starting calculating grain average Schmid factors.. 42 % Starting calculating grain average Schmid factors.. 43 % Starting calculating grain average Schmid factors.. 45 % Starting calculating grain average Schmid factors.. 47 % Starting calculating grain average Schmid factors.. 49 % Starting calculating grain average Schmid factors.. 51 % Starting calculating grain average Schmid factors.. 53 % Starting calculating grain average Schmid factors.. 55 % Starting calculating grain average Schmid factors.. 57 % Starting calculating grain average Schmid factors.. 58 % Starting calculating grain average Schmid factors.. 60 %
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. 42 % Starting calculating grain average Schmid factors.. 43 % Starting calculating grain average Schmid factors.. 45 % Starting calculating grain average Schmid factors.. 47 % Starting calculating grain average Schmid factors.. 49 % Starting calculating grain average Schmid factors.. 51 % Starting calculating grain average Schmid factors.. 53 % Starting calculating grain average Schmid factors.. 55 % Starting calculating grain average Schmid factors.. 57 % Starting calculating grain average Schmid factors.. 58 % Starting calculating grain average Schmid factors.. 60 %
Starting calculating grain average Schmid factors.. 62 % Starting calculating grain average Schmid factors.. 64 % Starting calculating grain average Schmid factors.. 66 % Starting calculating grain average Schmid factors.. 68 % Starting calculating grain average Schmid factors.. 70 % Starting calculating grain average Schmid factors.. 72 % Starting calculating grain average Schmid factors.. 74 % Starting calculating grain average Schmid factors.. 75 % Starting calculating grain average Schmid factors.. 77 %
</pre>
Starting calculating grain average Schmid factors.. 62 % Starting calculating grain average Schmid factors.. 64 % Starting calculating grain average Schmid factors.. 66 % Starting calculating grain average Schmid factors.. 68 % Starting calculating grain average Schmid factors.. 70 % Starting calculating grain average Schmid factors.. 72 % Starting calculating grain average Schmid factors.. 74 % Starting calculating grain average Schmid factors.. 75 % Starting calculating grain average Schmid factors.. 77 %
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. 62 % Starting calculating grain average Schmid factors.. 64 % Starting calculating grain average Schmid factors.. 66 % Starting calculating grain average Schmid factors.. 68 % Starting calculating grain average Schmid factors.. 70 % Starting calculating grain average Schmid factors.. 72 % Starting calculating grain average Schmid factors.. 74 % Starting calculating grain average Schmid factors.. 75 % Starting calculating grain average Schmid factors.. 77 %
Starting calculating grain average Schmid factors.. 79 % Starting calculating grain average Schmid factors.. 81 % Starting calculating grain average Schmid factors.. 83 % Starting calculating grain average Schmid factors.. 85 % Starting calculating grain average Schmid factors.. 87 % Starting calculating grain average Schmid factors.. 89 % Starting calculating grain average Schmid factors.. 91 % Starting calculating grain average Schmid factors.. 92 % Starting calculating grain average Schmid factors.. 94 % Starting calculating grain average Schmid factors.. 96 % Starting calculating grain average Schmid factors.. 98 % Starting calculating grain average Schmid factors.. 100 % Finished calculating grain average Schmid factors (0:00:01)
</pre>
Starting calculating grain average Schmid factors.. 79 % Starting calculating grain average Schmid factors.. 81 % Starting calculating grain average Schmid factors.. 83 % Starting calculating grain average Schmid factors.. 85 % Starting calculating grain average Schmid factors.. 87 % Starting calculating grain average Schmid factors.. 89 % Starting calculating grain average Schmid factors.. 91 % Starting calculating grain average Schmid factors.. 92 % Starting calculating grain average Schmid factors.. 94 % Starting calculating grain average Schmid factors.. 96 % Starting calculating grain average Schmid factors.. 98 % Starting calculating grain average Schmid factors.. 100 % Finished calculating grain average Schmid factors (0:00:01)
end{sphinxVerbatim}
Starting calculating grain average Schmid factors.. 79 % Starting calculating grain average Schmid factors.. 81 % Starting calculating grain average Schmid factors.. 83 % Starting calculating grain average Schmid factors.. 85 % Starting calculating grain average Schmid factors.. 87 % Starting calculating grain average Schmid factors.. 89 % Starting calculating grain average Schmid factors.. 91 % Starting calculating grain average Schmid factors.. 92 % Starting calculating grain average Schmid factors.. 94 % Starting calculating grain average Schmid factors.. 96 % Starting calculating grain average Schmid factors.. 98 % Starting calculating grain average Schmid factors.. 100 % Finished calculating grain average Schmid factors (0:00:01)
[16]:
ebsd_map.plot_average_grain_schmid_factors_map()
[16]:
<defdap.plotting.MapPlot at 0x7fe47f573ee0>
Single grain analysis¶
The locate_grain method allows interactive selection of a grain of intereset to apply any analysis to. Clicking on grains in the map will highlight the grain and print out the grain ID (position in the grain list) of the grain.
[17]:
ebsd_map.locate_grain()
[17]:
<defdap.plotting.MapPlot at 0x7fe47a6f29b0>
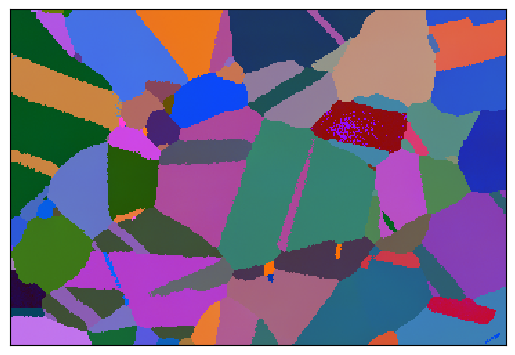
A built-in example is to calculate the average orientation of the grain and plot this orientation in a IPF
[18]:
grain_id = 48
grain = ebsd_map[grain_id]
grain.calc_average_ori() # stored as a quaternion named grain.refOri
print(grain.ref_ori)
grain.plot_ref_ori(direction=[0, 0, 1])
[0.3393, 0.1397, 0.4032, 0.8383]
[18]:
<defdap.plotting.PolePlot at 0x7fe47e9edd80>
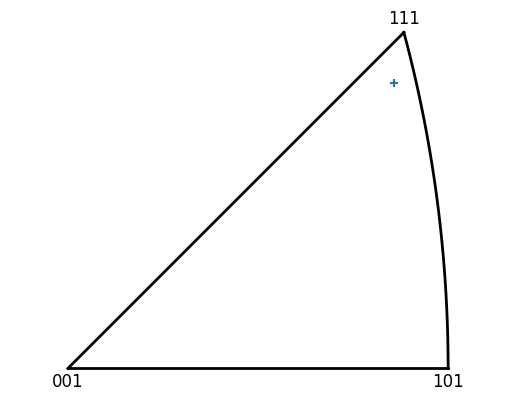
The spread of orientations in a given grain can also be plotted on an IPF
[19]:
plot = grain.plot_ori_spread(direction=np.array([0, 0, 1]), c='b', s=1, alpha=0.2)
grain.plot_ref_ori(direction=[0, 0, 1], c='k', plot=plot)
[19]:
<defdap.plotting.PolePlot at 0x7fe47f78d3c0>
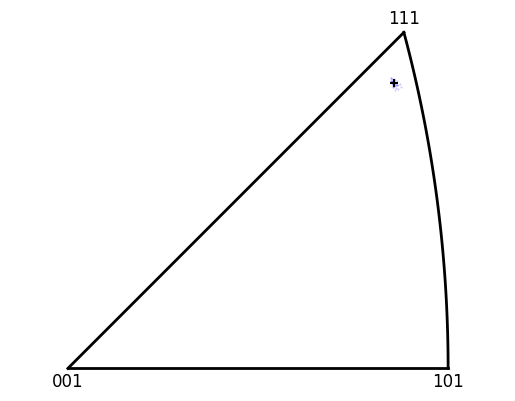
The unit cell for the average grain orientation can also be ploted
[20]:
grain.plot_unit_cell()
[20]:
<defdap.plotting.CrystalPlot at 0x7fe47a2e2410>
Printing a list of the slip plane indices, angle of slip plane intersection with the screen (defined as counter-clockwise from upwards), colour defined for the slip plane and also the slip directions and corresponding Schmid factors, is also built in
[21]:
grain.print_slip_traces()
(111) Colour: blue Angle: 55.77
[011̅] SF: 0.026
[1̅01] SF: 0.025
[11̅0] SF: 0.051
(111̅) Colour: green Angle: 89.67
[011] SF: 0.003
[1̅01̅] SF: 0.003
[11̅0] SF: 0.006
(1̅11) Colour: red Angle: 29.68
[011̅] SF: 0.404
[101] SF: 0.432
[1̅1̅0] SF: 0.027
(11̅1) Colour: white Angle: 152.39
[01̅1̅] SF: 0.381
[1̅01] SF: 0.410
[110] SF: 0.029
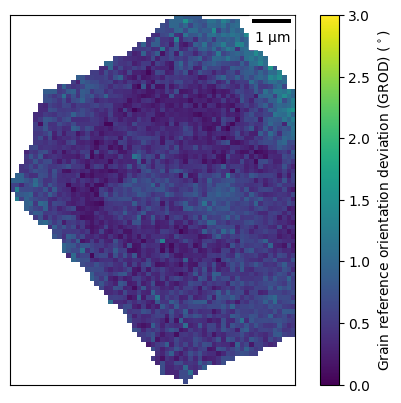
A second built-in example is to calcuate the grain misorientation, specifically the grain reference orientation deviation (GROD). This shows another feature of the locate_grain method, which stores the last selected grain in a variable called sel_grain in the EBSD map.
[22]:
if ebsd_map.sel_grain == None:
ebsd_map.sel_grain = ebsd_map[57]
ebsd_map.sel_grain.build_mis_ori_list()
ebsd_map.sel_grain.plot_mis_ori(plot_scale_bar=True, vmin=0, vmax=3)
[22]:
<defdap.plotting.GrainPlot at 0x7fe47a2e2d10>
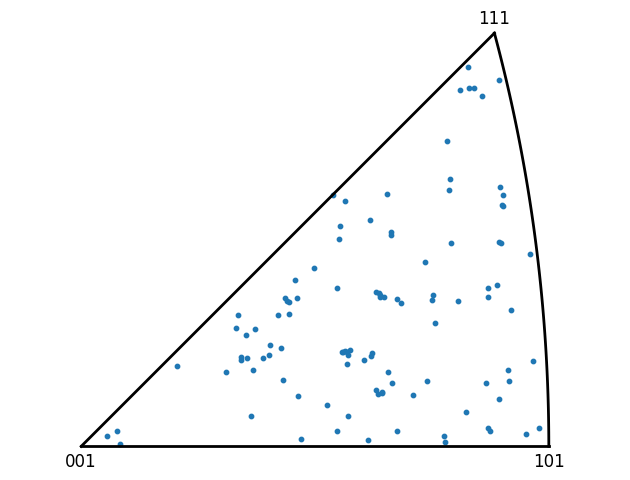
Multi grain analysis¶
Once an analysis routine has been prototyped for a single grain it can be applied to all the grains in a map using a loop over the grains and any results added to a list for use later. Of couse you could also apply to a smaller subset of grains as well.
[23]:
grain_av_oris = []
for grain in ebsd_map:
grain.calc_average_ori()
grain_av_oris.append(grain.ref_ori)
# Plot all the grain orientations in the map
Quat.plot_ipf(grain_av_oris, [0, 0, 1], ebsd_map.crystal_sym, marker='o', s=10)
plt.tight_layout()
Some common grain analysis routines are built into the EBSD map object, including:
[24]:
ebsd_map.calc_grain_av_oris()
Starting calculating grain mean orientations.. Starting calculating grain mean orientations.. 2 % Starting calculating grain mean orientations.. 4 % Starting calculating grain mean orientations.. 6 % Starting calculating grain mean orientations.. 8 % Starting calculating grain mean orientations.. 9 %
</pre>
Starting calculating grain mean orientations.. Starting calculating grain mean orientations.. 2 % Starting calculating grain mean orientations.. 4 % Starting calculating grain mean orientations.. 6 % Starting calculating grain mean orientations.. 8 % Starting calculating grain mean orientations.. 9 %
end{sphinxVerbatim}
Starting calculating grain mean orientations.. Starting calculating grain mean orientations.. 2 % Starting calculating grain mean orientations.. 4 % Starting calculating grain mean orientations.. 6 % Starting calculating grain mean orientations.. 8 % Starting calculating grain mean orientations.. 9 %
Starting calculating grain mean orientations.. 11 % Starting calculating grain mean orientations.. 13 % Starting calculating grain mean orientations.. 15 % Starting calculating grain mean orientations.. 17 % Starting calculating grain mean orientations.. 19 % Starting calculating grain mean orientations.. 21 % Starting calculating grain mean orientations.. 23 % Starting calculating grain mean orientations.. 25 % Starting calculating grain mean orientations.. 26 %
</pre>
Starting calculating grain mean orientations.. 11 % Starting calculating grain mean orientations.. 13 % Starting calculating grain mean orientations.. 15 % Starting calculating grain mean orientations.. 17 % Starting calculating grain mean orientations.. 19 % Starting calculating grain mean orientations.. 21 % Starting calculating grain mean orientations.. 23 % Starting calculating grain mean orientations.. 25 % Starting calculating grain mean orientations.. 26 %
end{sphinxVerbatim}
Starting calculating grain mean orientations.. 11 % Starting calculating grain mean orientations.. 13 % Starting calculating grain mean orientations.. 15 % Starting calculating grain mean orientations.. 17 % Starting calculating grain mean orientations.. 19 % Starting calculating grain mean orientations.. 21 % Starting calculating grain mean orientations.. 23 % Starting calculating grain mean orientations.. 25 % Starting calculating grain mean orientations.. 26 %
Starting calculating grain mean orientations.. 28 % Starting calculating grain mean orientations.. 30 % Starting calculating grain mean orientations.. 32 % Starting calculating grain mean orientations.. 34 % Starting calculating grain mean orientations.. 36 % Starting calculating grain mean orientations.. 38 % Starting calculating grain mean orientations.. 40 %
</pre>
Starting calculating grain mean orientations.. 28 % Starting calculating grain mean orientations.. 30 % Starting calculating grain mean orientations.. 32 % Starting calculating grain mean orientations.. 34 % Starting calculating grain mean orientations.. 36 % Starting calculating grain mean orientations.. 38 % Starting calculating grain mean orientations.. 40 %
end{sphinxVerbatim}
Starting calculating grain mean orientations.. 28 % Starting calculating grain mean orientations.. 30 % Starting calculating grain mean orientations.. 32 % Starting calculating grain mean orientations.. 34 % Starting calculating grain mean orientations.. 36 % Starting calculating grain mean orientations.. 38 % Starting calculating grain mean orientations.. 40 %
Starting calculating grain mean orientations.. 42 % Starting calculating grain mean orientations.. 43 % Starting calculating grain mean orientations.. 45 % Starting calculating grain mean orientations.. 47 % Starting calculating grain mean orientations.. 49 % Starting calculating grain mean orientations.. 51 % Starting calculating grain mean orientations.. 53 % Starting calculating grain mean orientations.. 55 % Starting calculating grain mean orientations.. 57 % Starting calculating grain mean orientations.. 58 % Starting calculating grain mean orientations.. 60 %
</pre>
Starting calculating grain mean orientations.. 42 % Starting calculating grain mean orientations.. 43 % Starting calculating grain mean orientations.. 45 % Starting calculating grain mean orientations.. 47 % Starting calculating grain mean orientations.. 49 % Starting calculating grain mean orientations.. 51 % Starting calculating grain mean orientations.. 53 % Starting calculating grain mean orientations.. 55 % Starting calculating grain mean orientations.. 57 % Starting calculating grain mean orientations.. 58 % Starting calculating grain mean orientations.. 60 %
end{sphinxVerbatim}
Starting calculating grain mean orientations.. 42 % Starting calculating grain mean orientations.. 43 % Starting calculating grain mean orientations.. 45 % Starting calculating grain mean orientations.. 47 % Starting calculating grain mean orientations.. 49 % Starting calculating grain mean orientations.. 51 % Starting calculating grain mean orientations.. 53 % Starting calculating grain mean orientations.. 55 % Starting calculating grain mean orientations.. 57 % Starting calculating grain mean orientations.. 58 % Starting calculating grain mean orientations.. 60 %
Starting calculating grain mean orientations.. 62 % Starting calculating grain mean orientations.. 64 % Starting calculating grain mean orientations.. 66 % Starting calculating grain mean orientations.. 68 % Starting calculating grain mean orientations.. 70 % Starting calculating grain mean orientations.. 72 % Starting calculating grain mean orientations.. 74 % Starting calculating grain mean orientations.. 75 % Starting calculating grain mean orientations.. 77 % Starting calculating grain mean orientations.. 79 % Starting calculating grain mean orientations.. 81 %
</pre>
Starting calculating grain mean orientations.. 62 % Starting calculating grain mean orientations.. 64 % Starting calculating grain mean orientations.. 66 % Starting calculating grain mean orientations.. 68 % Starting calculating grain mean orientations.. 70 % Starting calculating grain mean orientations.. 72 % Starting calculating grain mean orientations.. 74 % Starting calculating grain mean orientations.. 75 % Starting calculating grain mean orientations.. 77 % Starting calculating grain mean orientations.. 79 % Starting calculating grain mean orientations.. 81 %
end{sphinxVerbatim}
Starting calculating grain mean orientations.. 62 % Starting calculating grain mean orientations.. 64 % Starting calculating grain mean orientations.. 66 % Starting calculating grain mean orientations.. 68 % Starting calculating grain mean orientations.. 70 % Starting calculating grain mean orientations.. 72 % Starting calculating grain mean orientations.. 74 % Starting calculating grain mean orientations.. 75 % Starting calculating grain mean orientations.. 77 % Starting calculating grain mean orientations.. 79 % Starting calculating grain mean orientations.. 81 %
Starting calculating grain mean orientations.. 83 % Starting calculating grain mean orientations.. 85 % Starting calculating grain mean orientations.. 87 % Starting calculating grain mean orientations.. 89 % Starting calculating grain mean orientations.. 91 % Starting calculating grain mean orientations.. 92 % Starting calculating grain mean orientations.. 94 % Starting calculating grain mean orientations.. 96 % Starting calculating grain mean orientations.. 98 % Starting calculating grain mean orientations.. 100 % Finished calculating grain mean orientations (0:00:01)
</pre>
Starting calculating grain mean orientations.. 83 % Starting calculating grain mean orientations.. 85 % Starting calculating grain mean orientations.. 87 % Starting calculating grain mean orientations.. 89 % Starting calculating grain mean orientations.. 91 % Starting calculating grain mean orientations.. 92 % Starting calculating grain mean orientations.. 94 % Starting calculating grain mean orientations.. 96 % Starting calculating grain mean orientations.. 98 % Starting calculating grain mean orientations.. 100 % Finished calculating grain mean orientations (0:00:01)
end{sphinxVerbatim}
Starting calculating grain mean orientations.. 83 % Starting calculating grain mean orientations.. 85 % Starting calculating grain mean orientations.. 87 % Starting calculating grain mean orientations.. 89 % Starting calculating grain mean orientations.. 91 % Starting calculating grain mean orientations.. 92 % Starting calculating grain mean orientations.. 94 % Starting calculating grain mean orientations.. 96 % Starting calculating grain mean orientations.. 98 % Starting calculating grain mean orientations.. 100 % Finished calculating grain mean orientations (0:00:01)
[25]:
ebsd_map.calc_grain_mis_ori()
ebsd_map.plot_mis_ori_map(vmin=0, vmax=5, plot_gbs=True, plot_scale_bar=True)
Starting calculating grain misorientations.. Starting calculating grain misorientations.. 2 % Starting calculating grain misorientations.. 4 % Starting calculating grain misorientations.. 6 % Starting calculating grain misorientations.. 8 % Starting calculating grain misorientations.. 9 % Starting calculating grain misorientations.. 11 % Starting calculating grain misorientations.. 13 % Starting calculating grain misorientations.. 15 % Starting calculating grain misorientations.. 17 %
</pre>
Starting calculating grain misorientations.. Starting calculating grain misorientations.. 2 % Starting calculating grain misorientations.. 4 % Starting calculating grain misorientations.. 6 % Starting calculating grain misorientations.. 8 % Starting calculating grain misorientations.. 9 % Starting calculating grain misorientations.. 11 % Starting calculating grain misorientations.. 13 % Starting calculating grain misorientations.. 15 % Starting calculating grain misorientations.. 17 %
end{sphinxVerbatim}
Starting calculating grain misorientations.. Starting calculating grain misorientations.. 2 % Starting calculating grain misorientations.. 4 % Starting calculating grain misorientations.. 6 % Starting calculating grain misorientations.. 8 % Starting calculating grain misorientations.. 9 % Starting calculating grain misorientations.. 11 % Starting calculating grain misorientations.. 13 % Starting calculating grain misorientations.. 15 % Starting calculating grain misorientations.. 17 %
Starting calculating grain misorientations.. 19 % Starting calculating grain misorientations.. 21 % Starting calculating grain misorientations.. 23 % Starting calculating grain misorientations.. 25 % Starting calculating grain misorientations.. 26 % Starting calculating grain misorientations.. 28 % Starting calculating grain misorientations.. 30 % Starting calculating grain misorientations.. 32 % Starting calculating grain misorientations.. 34 % Starting calculating grain misorientations.. 36 % Starting calculating grain misorientations.. 38 % Starting calculating grain misorientations.. 40 %
</pre>
Starting calculating grain misorientations.. 19 % Starting calculating grain misorientations.. 21 % Starting calculating grain misorientations.. 23 % Starting calculating grain misorientations.. 25 % Starting calculating grain misorientations.. 26 % Starting calculating grain misorientations.. 28 % Starting calculating grain misorientations.. 30 % Starting calculating grain misorientations.. 32 % Starting calculating grain misorientations.. 34 % Starting calculating grain misorientations.. 36 % Starting calculating grain misorientations.. 38 % Starting calculating grain misorientations.. 40 %
end{sphinxVerbatim}
Starting calculating grain misorientations.. 19 % Starting calculating grain misorientations.. 21 % Starting calculating grain misorientations.. 23 % Starting calculating grain misorientations.. 25 % Starting calculating grain misorientations.. 26 % Starting calculating grain misorientations.. 28 % Starting calculating grain misorientations.. 30 % Starting calculating grain misorientations.. 32 % Starting calculating grain misorientations.. 34 % Starting calculating grain misorientations.. 36 % Starting calculating grain misorientations.. 38 % Starting calculating grain misorientations.. 40 %
Starting calculating grain misorientations.. 42 % Starting calculating grain misorientations.. 43 % Starting calculating grain misorientations.. 45 % Starting calculating grain misorientations.. 47 % Starting calculating grain misorientations.. 49 % Starting calculating grain misorientations.. 51 % Starting calculating grain misorientations.. 53 % Starting calculating grain misorientations.. 55 % Starting calculating grain misorientations.. 57 % Starting calculating grain misorientations.. 58 % Starting calculating grain misorientations.. 60 % Starting calculating grain misorientations.. 62 % Starting calculating grain misorientations.. 64 %
</pre>
Starting calculating grain misorientations.. 42 % Starting calculating grain misorientations.. 43 % Starting calculating grain misorientations.. 45 % Starting calculating grain misorientations.. 47 % Starting calculating grain misorientations.. 49 % Starting calculating grain misorientations.. 51 % Starting calculating grain misorientations.. 53 % Starting calculating grain misorientations.. 55 % Starting calculating grain misorientations.. 57 % Starting calculating grain misorientations.. 58 % Starting calculating grain misorientations.. 60 % Starting calculating grain misorientations.. 62 % Starting calculating grain misorientations.. 64 %
end{sphinxVerbatim}
Starting calculating grain misorientations.. 42 % Starting calculating grain misorientations.. 43 % Starting calculating grain misorientations.. 45 % Starting calculating grain misorientations.. 47 % Starting calculating grain misorientations.. 49 % Starting calculating grain misorientations.. 51 % Starting calculating grain misorientations.. 53 % Starting calculating grain misorientations.. 55 % Starting calculating grain misorientations.. 57 % Starting calculating grain misorientations.. 58 % Starting calculating grain misorientations.. 60 % Starting calculating grain misorientations.. 62 % Starting calculating grain misorientations.. 64 %
Starting calculating grain misorientations.. 66 % Starting calculating grain misorientations.. 68 % Starting calculating grain misorientations.. 70 % Starting calculating grain misorientations.. 72 % Starting calculating grain misorientations.. 74 % Starting calculating grain misorientations.. 75 % Starting calculating grain misorientations.. 77 % Starting calculating grain misorientations.. 79 % Starting calculating grain misorientations.. 81 % Starting calculating grain misorientations.. 83 % Starting calculating grain misorientations.. 85 % Starting calculating grain misorientations.. 87 % Starting calculating grain misorientations.. 89 % Starting calculating grain misorientations.. 91 % Starting calculating grain misorientations.. 92 % Starting calculating grain misorientations.. 94 % Starting calculating grain misorientations.. 96 % Starting calculating grain misorientations.. 98 %
</pre>
Starting calculating grain misorientations.. 66 % Starting calculating grain misorientations.. 68 % Starting calculating grain misorientations.. 70 % Starting calculating grain misorientations.. 72 % Starting calculating grain misorientations.. 74 % Starting calculating grain misorientations.. 75 % Starting calculating grain misorientations.. 77 % Starting calculating grain misorientations.. 79 % Starting calculating grain misorientations.. 81 % Starting calculating grain misorientations.. 83 % Starting calculating grain misorientations.. 85 % Starting calculating grain misorientations.. 87 % Starting calculating grain misorientations.. 89 % Starting calculating grain misorientations.. 91 % Starting calculating grain misorientations.. 92 % Starting calculating grain misorientations.. 94 % Starting calculating grain misorientations.. 96 % Starting calculating grain misorientations.. 98 %
end{sphinxVerbatim}
Starting calculating grain misorientations.. 66 % Starting calculating grain misorientations.. 68 % Starting calculating grain misorientations.. 70 % Starting calculating grain misorientations.. 72 % Starting calculating grain misorientations.. 74 % Starting calculating grain misorientations.. 75 % Starting calculating grain misorientations.. 77 % Starting calculating grain misorientations.. 79 % Starting calculating grain misorientations.. 81 % Starting calculating grain misorientations.. 83 % Starting calculating grain misorientations.. 85 % Starting calculating grain misorientations.. 87 % Starting calculating grain misorientations.. 89 % Starting calculating grain misorientations.. 91 % Starting calculating grain misorientations.. 92 % Starting calculating grain misorientations.. 94 % Starting calculating grain misorientations.. 96 % Starting calculating grain misorientations.. 98 %
Starting calculating grain misorientations.. 100 % Finished calculating grain misorientations (0:00:00)
</pre>
Starting calculating grain misorientations.. 100 % Finished calculating grain misorientations (0:00:00)
end{sphinxVerbatim}
Starting calculating grain misorientations.. 100 % Finished calculating grain misorientations (0:00:00)
[25]:
<defdap.plotting.MapPlot at 0x7fe47e9eff40>

There are also methods for plotting GND density, phases and boundaries. All of the plotting functions in DefDAP use the same parameters to modify the plot, examples seen so far are plot_gbs, plotScaleBar, vmin, vmax.
Linking the HRDIC and EBSD¶
Define homologous points¶
To register the two datasets, homologous points (points at the same material location) within each map are used to estimate a transformation between the two frames the data are defined in. The homologous points are selected manually using an interactive tool within DefDAP. To select homologous call the method setHomogPoint on each of the data maps, which will open a plot window with a button labelled ‘save point’ in the bottom right. You select a point by right clicking on the map, adjust
the position with the arrow and accept the point by with the save point button. Then select the same location in the other map. Note that as we set the location of the pattern image for the HRDIC map that the points can be selected on the pattern image rather than the strain data.
[26]:
dic_map.set_homog_point(map_name="pattern")
Loading img
[27]:
ebsd_map.set_homog_point()
The points are stored as a list of tuples (x, y) in each of the maps. This means the points can be set from previous values.
[28]:
dic_map.frame.homog_points
[28]:
[]
[29]:
ebsd_map.frame.homog_points
[29]:
[]
Here are some example homologous points for this data, after setting these by running the cells below you can view the locations in the maps by running the setHomogPoint methods (above) again
[30]:
dic_map.frame.homog_points = [
(36, 72),
(279, 27),
(162, 174),
(60, 157)
]
[31]:
ebsd_map.frame.homog_points = [
(68, 95),
(308, 45),
(191, 187),
(89, 174)
]
Link the maps¶
Finally the two data maps are linked. The type of transform between the two frames can be affine, projective, polynomial.
[32]:
dic_map.link_ebsd_map(ebsd_map, transform_type="affine")
# dic_map.link_ebsd_map(ebsd_map, transform_type="projective")
# dic_map.link_ebsd_map(ebsd_map, transform_type="polynomial", order=2)
Show the transformation¶
[33]:
from skimage import transform as tf
data = np.zeros((2000, 2000), dtype=float)
data[500:1500, 500:1500] = 1.
transform = dic_map.experiment.get_frame_transform(dic_map.frame, ebsd_map.frame)
dataWarped = tf.warp(data, transform)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(8,4))
ax1.set_title('Reference')
ax1.imshow(data)
ax2.set_title('Transformed')
ax2.imshow(dataWarped)
[33]:
<matplotlib.image.AxesImage at 0x7fe479de07c0>
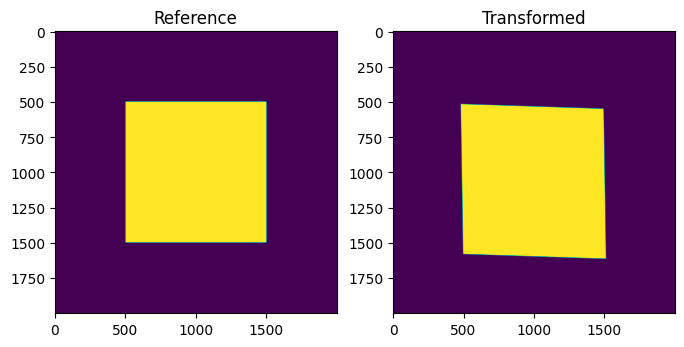
Segment into grains¶
The HRDIC map can now be segmented into grains using the grain boundaries detected in the EBSD map. Analysis rountines can then be applied to individual grain, as with the EBSD grains. The grain finding process will also attempt to link the grains between the EBSD and HRDIC and each grain in the HRDIC has a reference (ebsdGrain) to the corrosponding grain in the EBSD map.
[34]:
dic_map.data.generate('grains', min_grain_size=10)
- Starting finding grains..
</pre>
- Starting finding grains..
end{sphinxVerbatim}
Starting finding grains..
- Finished finding grains (0:00:00)
</pre>
- Finished finding grains (0:00:00)
end{sphinxVerbatim}
Finished finding grains (0:00:00)
[35]:
dic_map.plot_map('max_shear', vmin=0, vmax=0.10, plot_scale_bar=True, plot_gbs='pixel')
[35]:
<defdap.plotting.MapPlot at 0x7fe47a171750>
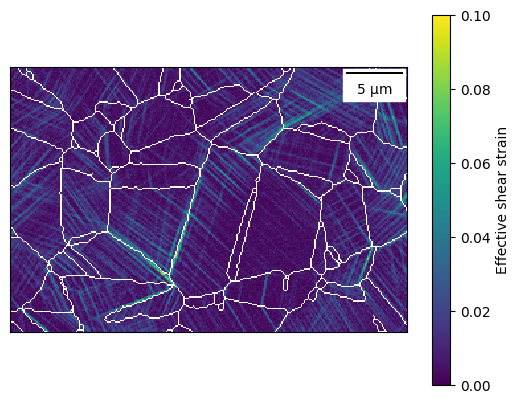
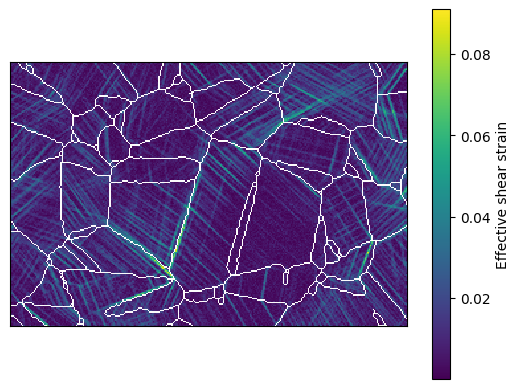
Now, a grain can also be selected interactively in the DIC map, in the same way a grain can be selected from an EBSD map. If displaySelected is set to true, then a pop-out window shows the map segmented for the grain
[36]:
dic_map.locate_grain(display_grain=True)
# This produces an error after a 3rd grain is
# selected related to removing the colourbar
# from the previous state. Needs looking at.
[36]:
<defdap.plotting.MapPlot at 0x7fe4797ccc40>
Plotting examples¶
Some of the plotting features are shown in examples below.
Built-in plots¶
[37]:
plot = dic_map.plot_map(
'max_shear', vmin=0, vmax=0.10,
plot_scale_bar=True, plot_gbs='line'
)
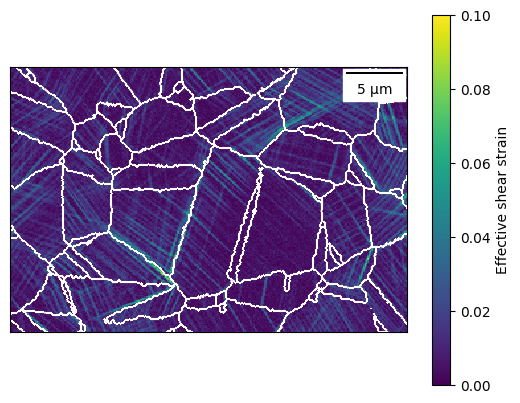
[38]:
plot = ebsd_map.plot_euler_map(
plot_scale_bar=True, plot_gbs=True,
highlight_grains=[10, 20, 45], highlight_alpha=0.9, highlight_colours=['y']
)
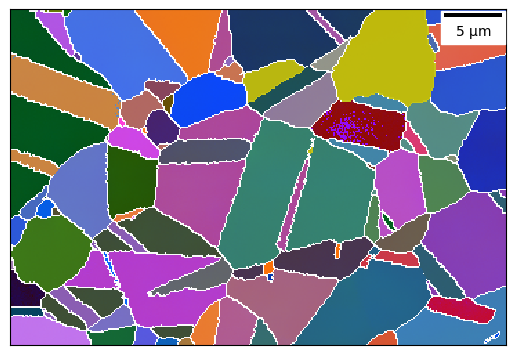
[39]:
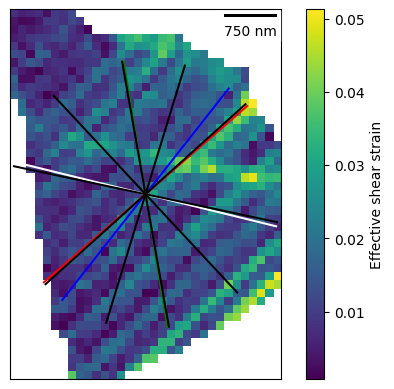
dic_grain_id = 42
dic_grain = dic_map[dic_grain_id]
plot = dic_grain.plot_max_shear(
plot_scale_bar=True, plot_slip_traces=True, plot_slip_bands=True
)
Number of bands detected: 5
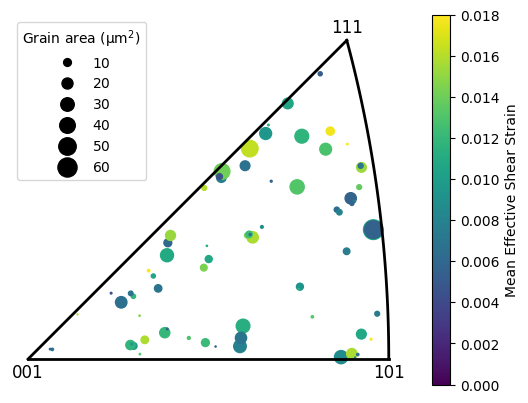
IPF plotting¶
This plot will show the positions of selected grains in an IPF pole figure, with the marker size representing grain area and mean effective shear strain.
[40]:
# For all grains in the DIC map
# Make an array of quaternions
grain_oris = [grain.ebsd_grain.ref_ori for grain in dic_map]
# Make an array of grain area
grain_areas = np.array([len(grain) for grain in dic_map]) * dic_map.scale**2
# Scaling the grain area, so that the maximum size of a marker is 200 points^2
grain_area_scaling = 200. / grain_areas.max()
# Make an array of mean effective shear strain
grain_strains = [np.array(grain.data.max_shear).mean() for grain in dic_map]
plot = Quat.plot_ipf(grain_oris, direction=[1,0,0], sym_group='cubic', marker='o',
s=grain_areas*grain_area_scaling, vmin=0, vmax=0.018, cmap='viridis', c=grain_strains)
plot.add_colour_bar(label='Mean Effective Shear Strain')
plot.add_legend(scaling=grain_area_scaling)
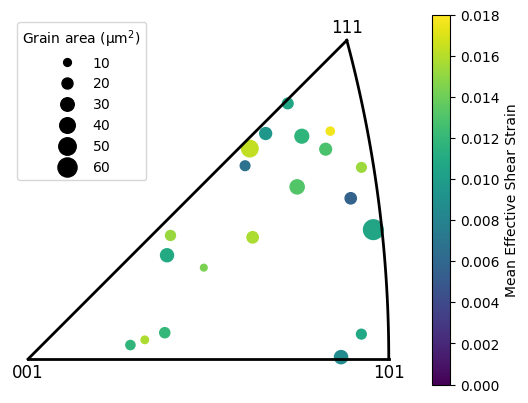
[41]:
# For selected grains in the DIC map
# Select grains from the DIC map
dic_grain_ids = [2, 5, 7, 9, 15, 17, 18, 23, 29, 32, 33, 37, 40, 42, 49, 50, 51, 54, 58, 60]
# Make an array of quaternions
grain_oris = np.array([dic_map[grain_id].ebsd_grain.ref_ori for grain_id in dic_grain_ids])
# Make an array of grain area
grain_areas = np.array([len(dic_map[grain_id]) for grain_id in dic_grain_ids]) * dic_map.scale**2
# Scaling the grain area, so that the maximum size of a marker is 200 points^2
grain_area_scaling = 200. / grain_areas.max()
# Make an array of mean effective shear strain
grain_strains = np.array([np.mean(dic_map[grain].data.max_shear) for grain in dic_grain_ids])
plot = Quat.plot_ipf(grain_oris, direction=[1,0,0], sym_group='cubic', marker='o',
s=grain_areas*grain_area_scaling, vmin=0, vmax=0.018, cmap='viridis', c=grain_strains)
plot.add_colour_bar(label='Mean Effective Shear Strain')
plot.add_legend(scaling=grain_area_scaling)
Create your own¶
[42]:
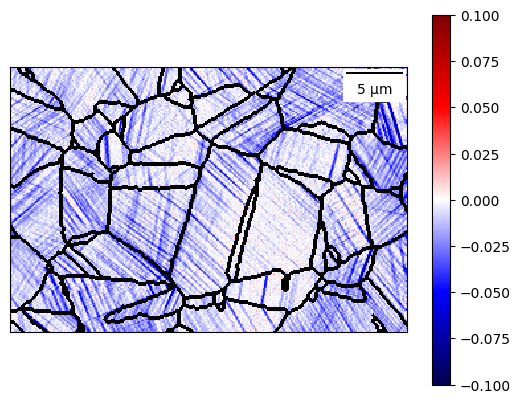
from defdap.plotting import MapPlot, GrainPlot, HistPlot
[43]:
map_data = dic_map.data['e'][0,0]
map_data = dic_map.crop(map_data)
plot = MapPlot.create(
dic_map, map_data,
vmin=-0.1, vmax=0.1, plot_colour_bar=True, cmap="seismic",
plot_gbs=True, dilate_boundaries=True, boundary_colour='black'
)
plot.add_scale_bar()
Functions for grain averaging and grain segmentation¶
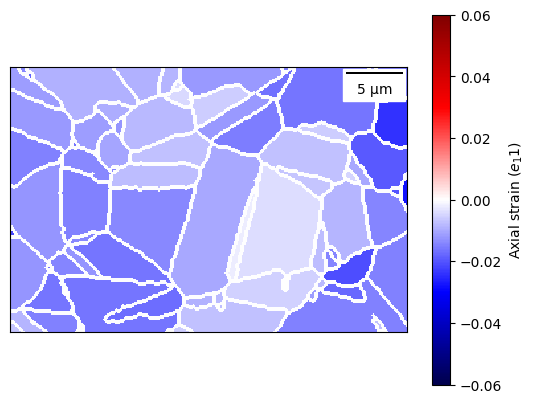
[44]:
plot = dic_map.plot_grain_data_map(
map_data,
vmin=-0.06, vmax=0.06, plot_colour_bar=True,
cmap="seismic", clabel="Axial strain ($e_11$)",
plot_scale_bar=True
)
plot.add_grain_boundaries(dilate=True, colour="white")
[44]:
<matplotlib.image.AxesImage at 0x7fe4791e7970>
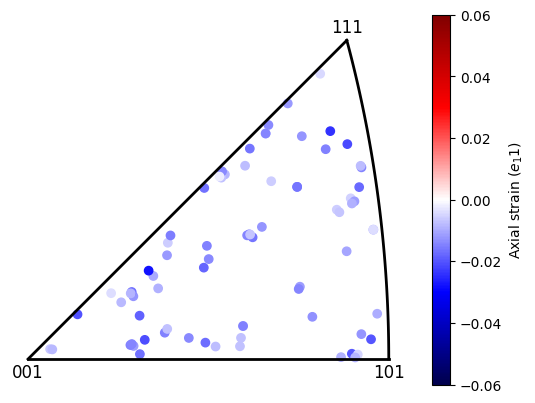
[45]:
plot = dic_map.plot_grain_data_ipf(
np.array((1,0,0)), map_data, marker='o',
vmin=-0.06, vmax=0.06, plot_colour_bar=True,
clabel="Axial strain ($e_11$)", cmap="seismic",
)
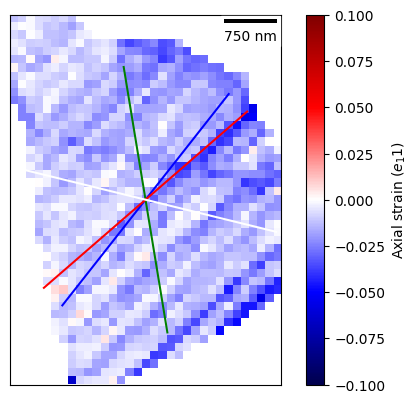
[46]:
dic_grain_id = 42
dic_grain = dic_map[dic_grain_id]
plot = dic_grain.plot_grain_data(
map_data,
vmin=-0.1, vmax=0.1, plot_colour_bar=True,
clabel="Axial strain ($e_11$)", cmap="seismic",
plot_scale_bar=True
)
plot.add_slip_traces()
Composite plots¶
By utilising some additional functionality within matplotlib, composite plots can be produced.
[47]:
from matplotlib import gridspec
[48]:
# Create a figure with 3 sets of axes
fig = plt.figure(figsize=(8, 4))
gs = gridspec.GridSpec(2, 2, width_ratios=[3, 1],
wspace=0.15, hspace=0.15,
left=0.02, right=0.98,
bottom=0.12, top=0.95)
ax0 = plt.subplot(gs[:, 0])
ax1 = plt.subplot(gs[0, 1])
ax2 = plt.subplot(gs[1, 1])
# add a strain map
plot0 = dic_map.plot_map(
map_name='max_shear',
ax=ax0, fig=fig,
vmin=0, vmax=0.08, plot_scale_bar=True,
plot_gbs=True, dilate_boundaries=True
)
# add an IPF of grain orientations
dic_oris = []
for grain in dic_map:
if len(grain) > 20:
dic_oris.append(grain.ref_ori)
plot1 = Quat.plot_ipf(
dic_oris, np.array((1,0,0)), 'cubic',
ax=ax1, fig=fig, s=10
)
# add histrogram of strain values
plot2 = HistPlot.create(
dic_map.crop(dic_map.data.max_shear),
ax=ax2, fig=fig, marker='o', markersize=2,
axes_type="logy", bins=50, range=(0,0.06)
)
plot2.ax.set_xlabel("Effective shear strain")
/home/docs/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/plotting.py:1390: UserWarning: marker is redundantly defined by the 'marker' keyword argument and the fmt string "o" (-> marker='o'). The keyword argument will take precedence.
self.ax.plot(x_vals, y_vals, line, label=label, **kwargs)
[48]:
Text(0.5, 18.72222222222218, 'Effective shear strain')
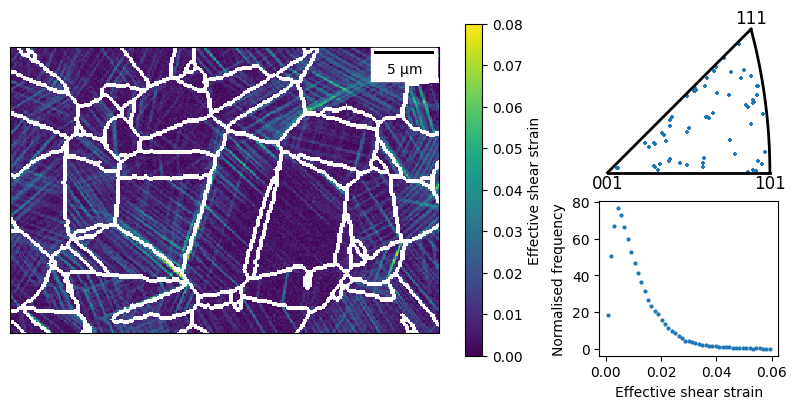
Figures can be saved to raster (png, jpg, ..) and vector formats (eps, svg), the format is guessed from the file extension given. The last displayed figure can be saved using:
[49]:
#plt.savefig("test_save_fig.png", dpi=200)
#plt.savefig("test_save_fig.eps", dpi=200)
[50]:
fig, ((ax0, ax1), (ax2, ax3)) = plt.subplots(2, 2, figsize=(8, 6))
dic_grain_id = 42
dic_grain = dic_map[dic_grain_id]
# add a strain map
plot0 = dic_grain.plot_max_shear(
ax=ax0, fig=fig,
vmin=0, vmax=0.08, plot_scale_bar=True,
plot_slip_traces=True
)
# add a misorientation
ebsd_grain = dic_grain.ebsd_grain
plot1 = ebsd_grain.plot_mis_ori(component=0, ax=ax1, fig=fig, vmin=0, vmax=1, clabel="GROD", plot_scale_bar=True)
# add an IPF
plot2 = ebsd_grain.plot_ori_spread(
direction=np.array((1,0,0)), c='b', s=1, alpha=0.2,
ax=ax2, fig=fig
)
ebsd_grain.plot_ref_ori(
direction=np.array((1,0,0)), c='k', s=100, plot=plot2
)
# add histrogram of strain values
plot3 = HistPlot.create(
dic_map.crop(dic_map.data.max_shear),
ax=ax3, fig=fig,
axes_type="logy", bins=50, range=(0,0.06))
plot3.ax.set_xlabel("Effective shear strain")
plt.tight_layout()
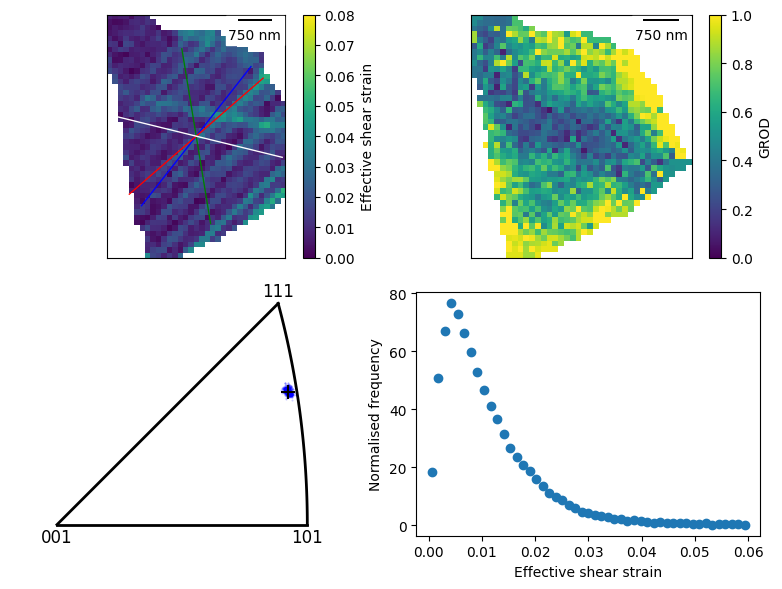
[ ]:
[ ]:
[ ]:
[51]:
import numpy as np
import matplotlib.pyplot as plt
import defdap.hrdic as hrdic
import defdap.ebsd as ebsd
from defdap.quat import Quat
from defdap.experiment import Experiment
[52]:
dic_path = "../../tests/data/"
dic_map = hrdic.Map(dic_path, "testDataDIC.txt")
ebsd_path = "../../tests/data/testDataEBSD"
ebsd_map = ebsd.Map(ebsd_path, dataType = 'OxfordBinary')
- Starting loading HRDIC data..
</pre>
- Starting loading HRDIC data..
end{sphinxVerbatim}
Starting loading HRDIC data..
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/file_readers.py:642, in DICDataLoader.get_loader(data_type)
641 try:
--> 642 loader = {
643 'davis': DavisLoader,
644 'openpiv': OpenPivLoader,
645 }[data_type]
646 except KeyError:
KeyError: 'testdatadic.txt'
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
Cell In[52], line 2
1 dic_path = "../../tests/data/"
----> 2 dic_map = hrdic.Map(dic_path, "testDataDIC.txt")
4 ebsd_path = "../../tests/data/testDataEBSD"
5 ebsd_map = ebsd.Map(ebsd_path, dataType = 'OxfordBinary')
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/hrdic.py:111, in Map.__init__(self, *args, **kwargs)
108 self.ydim = None # size of map along y (from header)
110 # Call base class constructor
--> 111 super(Map, self).__init__(*args, **kwargs)
113 self.corr_val = None # correlation value
115 self.ebsd_map = None # EBSD map linked to DIC map
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/base.py:87, in Map.__init__(self, file_name, data_type, experiment, increment, frame, map_name)
84 self.profile_plot = None
86 self.file_name = Path(file_name)
---> 87 self.load_data(self.file_name, data_type=data_type)
89 self.data.add_generator(
90 'proxigram', self.calc_proxigram, unit='', type='map', order=0,
91 cropped=True
92 )
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/utils.py:48, in report_progress.<locals>.decorator.<locals>.wrapper(*args, **kwargs)
46 try:
47 while True:
---> 48 prog = next(generator)
49 if type(prog) is str:
50 printFinal = False
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/hrdic.py:199, in Map.load_data(self, file_name, data_type)
187 @report_progress("loading HRDIC data")
188 def load_data(self, file_name, data_type=None):
189 """Load DIC data from file.
190
191 Parameters
(...)
197
198 """
--> 199 data_loader = DICDataLoader.get_loader(data_type)
200 data_loader.load(file_name)
202 metadata_dict = data_loader.loaded_metadata
File ~/checkouts/readthedocs.org/user_builds/defdap/envs/develop/lib/python3.10/site-packages/defdap/file_readers.py:647, in DICDataLoader.get_loader(data_type)
642 loader = {
643 'davis': DavisLoader,
644 'openpiv': OpenPivLoader,
645 }[data_type]
646 except KeyError:
--> 647 raise ValueError(f"No loader for DIC data of type {data_type}.")
648 return loader()
ValueError: No loader for DIC data of type testdatadic.txt.
[53]:
my_experiment = Experiment()
inc = my_experiment.add_increment(order=1)
inc.add_map('dic', dic_map)
inc.add_map('ebsd', ebsd_map)
[ ]:
[ ]:
[ ]: